- E-mail:support@braincase.cn
- Tel:18971216876
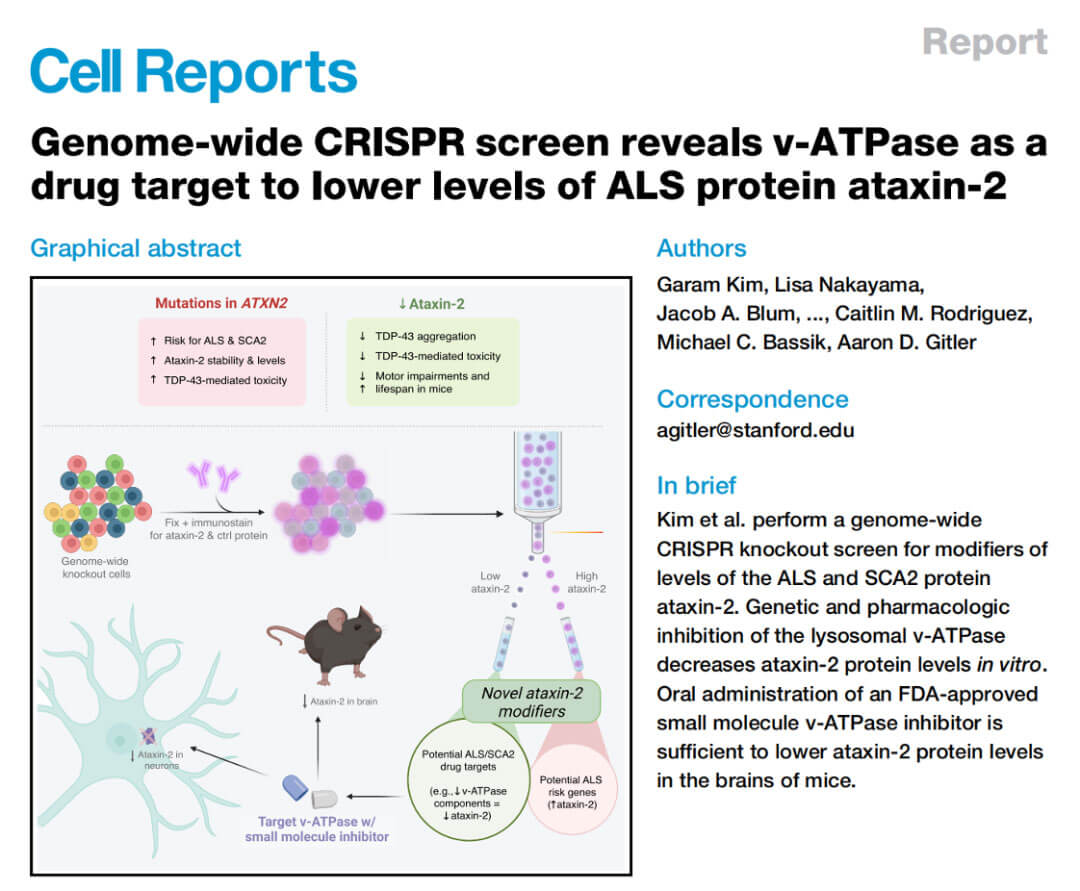
In October 2022, Professor Aaron D. Gitler of Stanford University published the research work "Genome-wide CRISPR screen reveals v-ATPase as a drug target to lower levels of ALS protein ataxin-2" in Cell Reports. This study used genome-wide CRISPR Knockout combined with flow cytometric sorting technology (FACS) screened and found multiple key genes that regulate ataxia gene 2 (ATXN2) protein levels, and verified through a series of in vivo and in vitro experiments that the screening results encoded lysosomal vesicles. Genes for components of the vesicular ATPase (v-ATPase) regulate endogenous ATXN2 protein levels. Multiple FDA-approved small molecule inhibitors of v-ATPase effectively reduce ATXN2 protein levels in mouse and human neurons, and oral administration of one of these drugs, such as etidronate, reduces ATXN2 protein in mouse brains level.
Ataxin-2 (ATXN2) is an RNA-binding protein and a regulator of stress granule assembly. It is related to the pathogenesis of various neurodegenerative diseases. Among them, the CAG trinucleotide repeat expansion copy of exon 1 of the ATXN2 gene A count >32 times is the cause of spinocerebellar ataxia type 2 (SCA2). Studies have shown that the intermediate repeat expansion of the ATXN2 gene that does not reach the SCA2 threshold is a risk factor for amyotrophic lateral sclerosis (ALS). Some studies have pointed out that ALS patients carrying intermediate repeat expansions of the ATXN2 gene have an earlier age of onset and shorter survival time.
Currently, a therapeutic strategy using antisense oligonucleotides to target ATXN2 to reduce its protein levels has entered human clinical trials. Developing new, safer and cheaper treatments to reduce ATXN2 protein levels can provide patients with more treatment options and reduce adverse reactions. At the same time, it can also help us understand the normal regulatory process of ATXN2 protein.
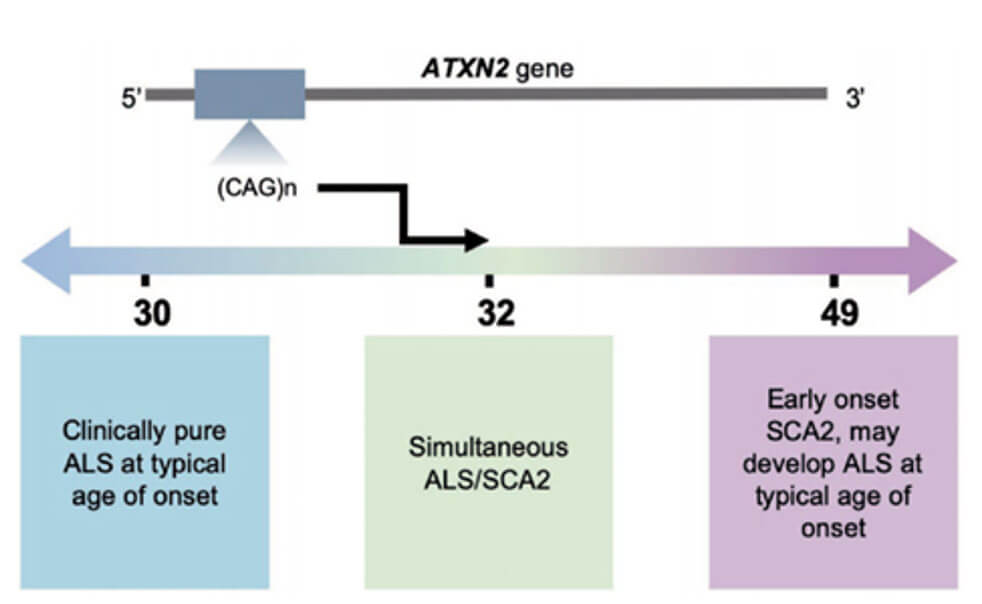
Temporal changes in ALS symptoms relative to SCA2 symptoms depend on the length of the ATXN2 expansion
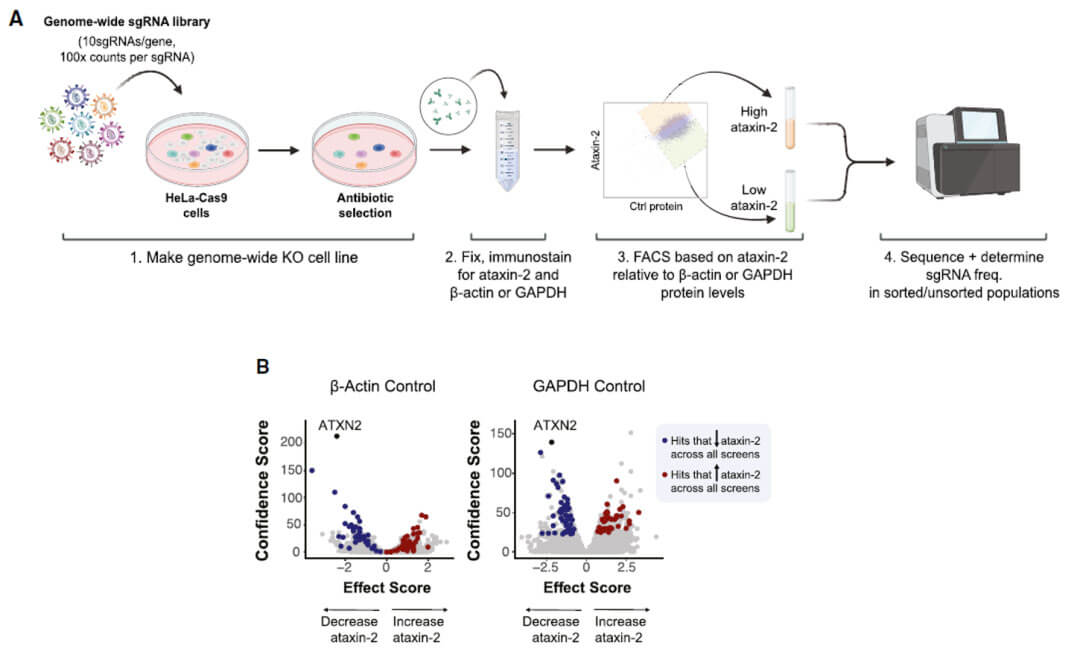
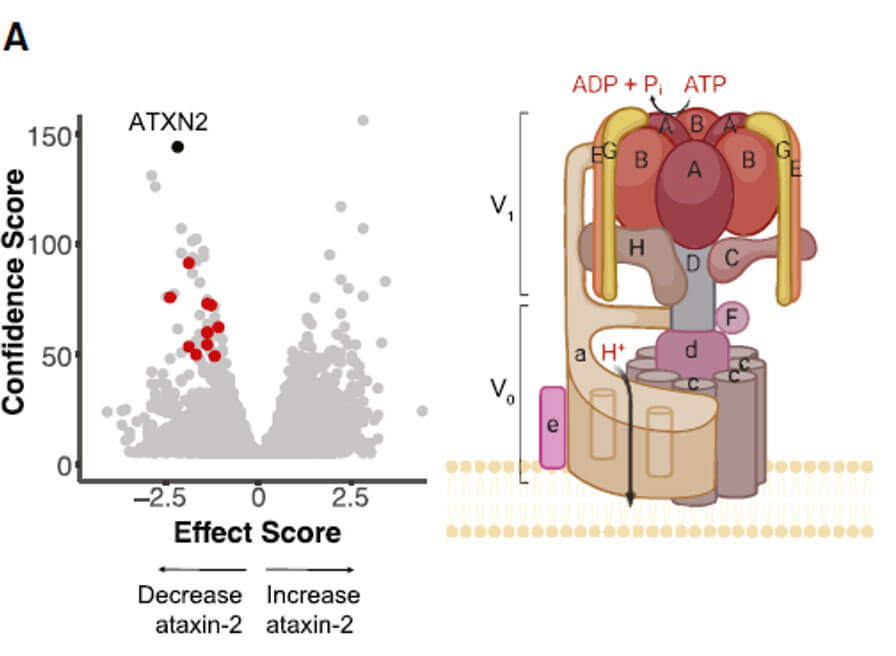
The authors used a genome-wide screening method combining CRISPR-Cas9 and FACS to sort out cell libraries with significantly up- and down-regulation of ATXN2 protein levels. They performed second-generation sequencing on this part of the cell library and found that factors affecting the endogenous protein levels of ATXN2 were found. candidate genes. The genes selected in both experiments are shown in the volcano plot in blue (down-regulated ATXN2 protein level) and red (up-regulated ATXN2 protein level) respectively.
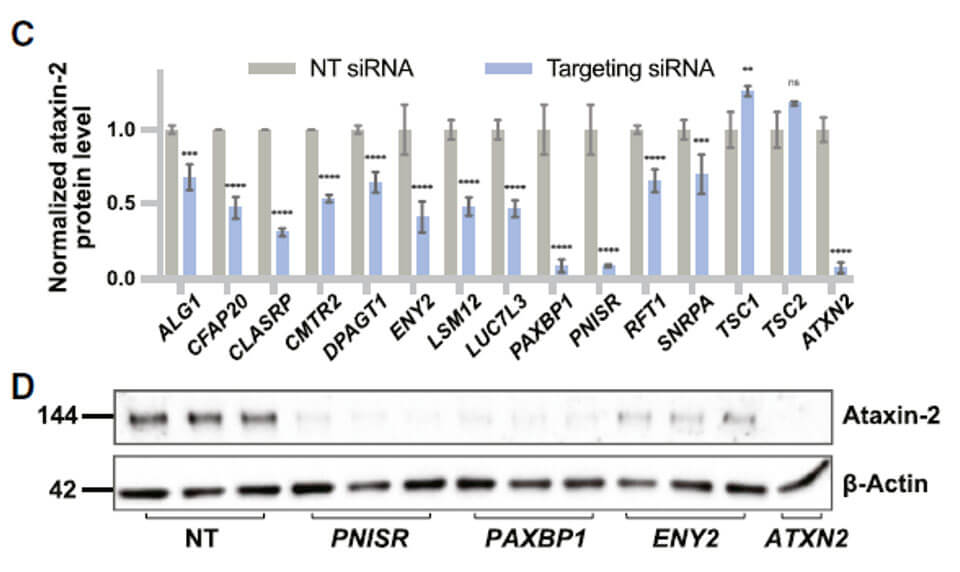
In order to further verify the feasibility of the screening system, the authors used siRNA to knock down the selected candidate genes and detect changes in ATXN2 protein levels. Verify that the selected candidate genes can indeed affect the protein level of ATXN2.
The selected candidate genes were classified according to function and subcellular localization. The authors found that knocking out a series of v-ATPases (ATP6V0C, ATP6V1A, ATP6V1B2, ATP6V1C1, ATP6V1D) will reduce the protein level of ATXN2. And it was verified at the protein level through siRNA knockdown. The knockdown of v-ATPase only affects the protein level of ATXN2 but does not affect the mRNA level, implying that the knockdown of v-ATPase affects the degradation of ATXN2 protein. . (The authors do not explain whether degradation occurs via proteasomes or lysosomes)
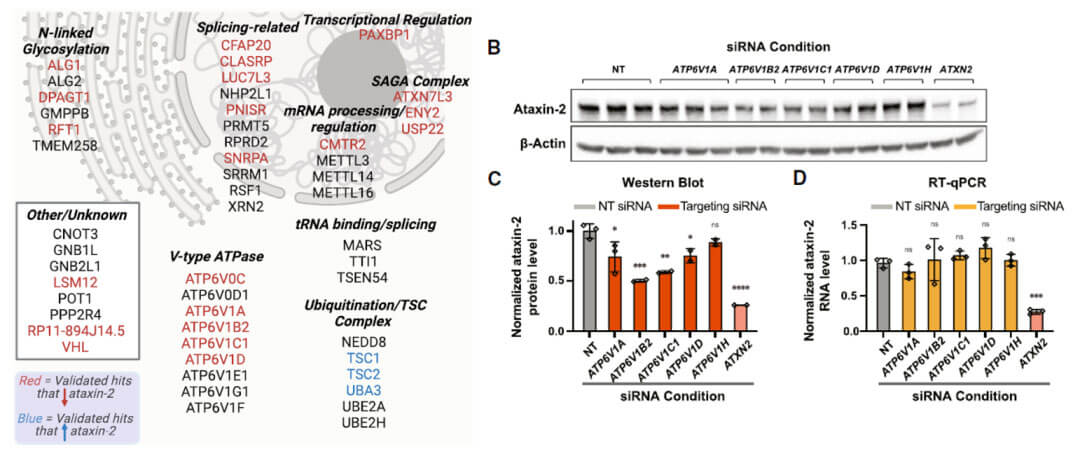
v-ATPase is an ATP hydrolysis-driven proton pump found in the intracellular membrane system of eukaryotes. It mainly uses the energy generated by hydrolyzing ATP to maintain hydrogen ion transport, thereby generating a transmembrane electrochemical gradient and regulating the pH value inside and outside the biological membrane. Abnormal functions of different subunits can lead to a variety of disease types. For example, mutations in ATP6V0A4 and ATP6V1B1 genes cause distal renal tubular acidosis. Mutations in ATP6V1C1, ATP6V1B2, ATP6V0A3, and ATP6V0D2 genes lead to diseases of bone development and metabolism. ATP6V1C1 plays an important role in breast cancer. play a certain role in the pathogenesis of Studies have also shown that ATP6V0A4 and ATP6V1B2 gene mutations are often associated with severe sensorineural deafness. With the deepening of neurobiological research, the role of v-ATPase in neurological diseases has gradually attracted attention.
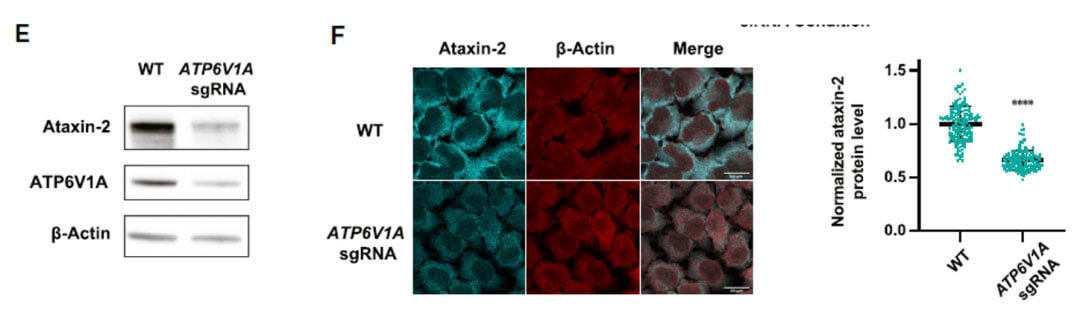
CRISPR-Cas9 technology was used to construct an ATP6V1A knockout cell line. By detecting protein levels and immunofluorescence, ATP6V1A knockout indeed reduced the protein level of ATXN2.
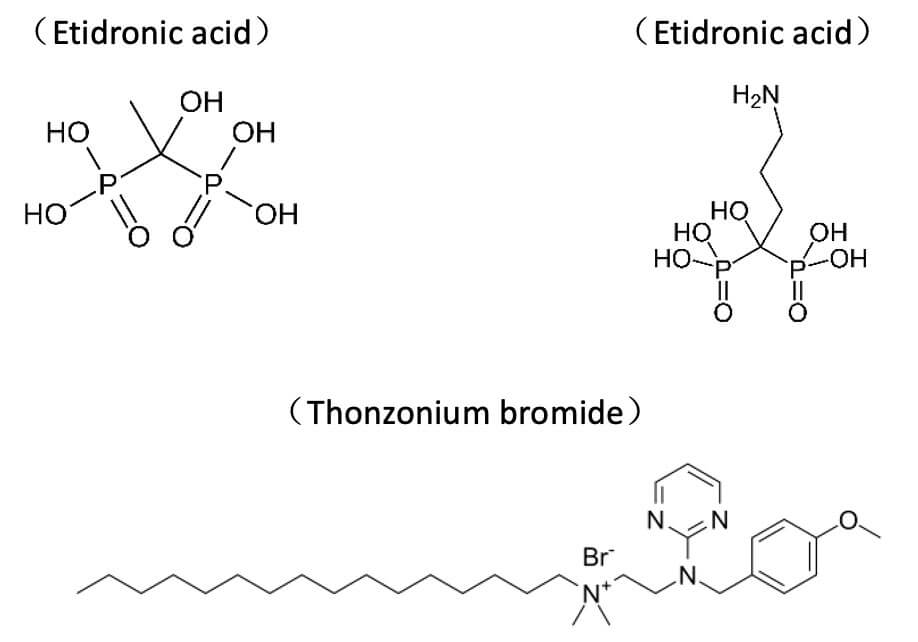
In order to find small molecule inhibitors that can affect ATXN2 protein levels by regulating the function of v-ATPase, the authors found three FDA-approved drugs (Etidronate, Alendronate and Thonzonium). The first two are bisphosphonates, which are combined with inorganic pyrophosphate. The similarity leads to its ability to inhibit the activity of v-ATPase, which exerts its inhibitory effect by uncoupling the proton transport of the v-ATPase proton pump and ATPase activity). Due to its good safety profile, Etidronate is recommended by the author. It conducted further research.
Treatment of primary neuronal cells derived from two different methods with etidronate can significantly down-regulate the protein level of ATXN2, with a significant dose-dependent effect.
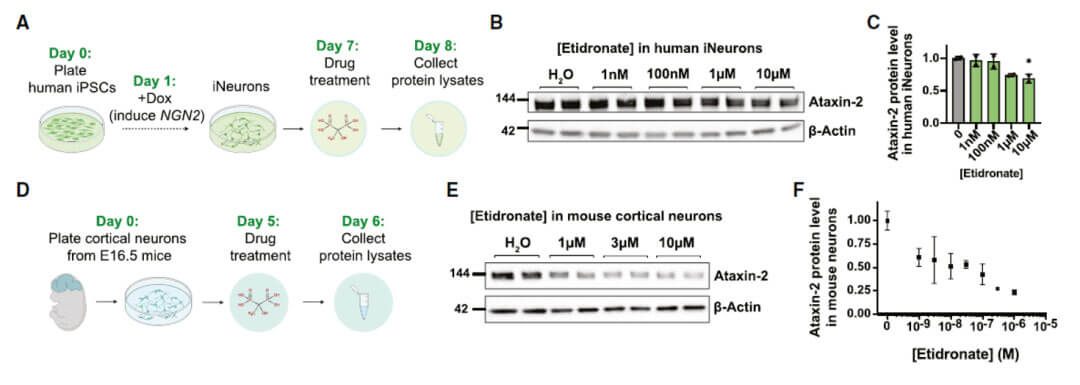
Treatment with etidronate also alleviates arsenite-induced stress granule formation.
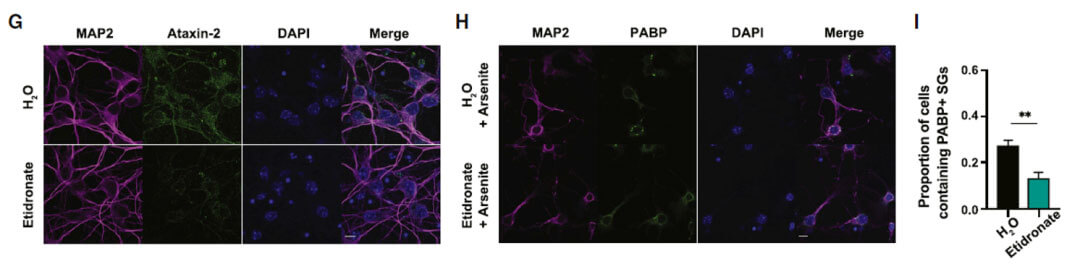
Etidronate can significantly reduce the protein level of ATXN2 in the cerebral cortex of mice after one week of oral administration.
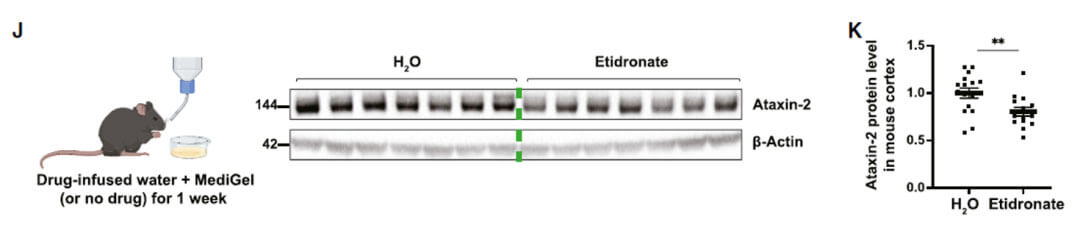
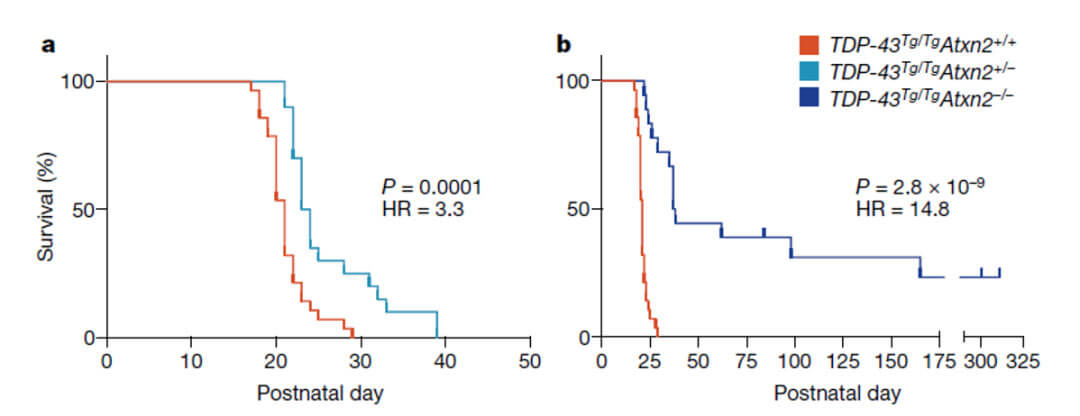
As the authors mentioned in the Discussion section, this study still has certain limitations. Although a decrease in ATXN2 protein levels was observed after oral administration of etidronate to mice, whether Etidronate can alleviate the corresponding movement disorders and neurodegeneration-related phenotypes Still not sure. Because the author found in another article that the corresponding mouse model (TDP-43Tg/Tg) died before being able to eat independently, the use of etidronate was restricted. However, since Etidronate can effectively reduce the protein level of ATXN2 in the body, the authors believe that Etidronate can play a role as a new therapeutic drug in ALS or SCA2.
In summary, this article proposes that v-ATPase is a drug target for amyotrophic lateral sclerosis and spinocerebellar ataxia type 2, and demonstrates the utility of CRISPR-Cas9 and FACS-based screening systems in identifying disease-related proteins and drug development. The value and research ideas are worthy of reference and learning.