- E-mail:BD@ebraincase.com
- Tel:+8618971215294
RNA interference (RNA interference, abbreviated as RNAi) refers to a gene silencing phenomenon induced by double-stranded RNA in molecular biology, and its mechanism is to inhibit gene expression by hindering the transcription or translation of specific genes. When double-stranded RNA homologous to the coding region of endogenous mRNA is introduced into cells, the mRNA is degraded and gene expression is silenced. Figure 1.
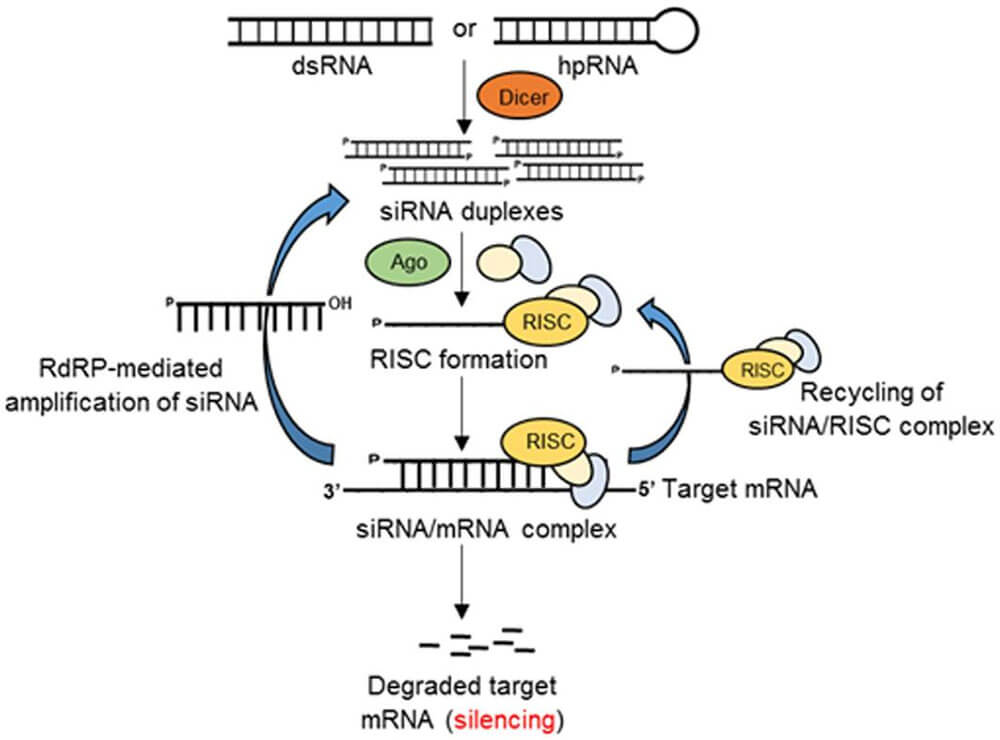
Figure 1. Schematic diagram of RNAi (Source: Majumdar Rajtilak et al., Frontiers in Plant Science, 2017)
Since RNAi has a high degree of sequence specificity, it can specifically silence genes, thereby obtaining loss of gene function or reduction of gene expression. Therefore, it can be used as a powerful tool for functional genomics research. In practical applications, the transcription of siRNA (small interfering RNA) mediated by viral vectors for RNA interference has matured, effectively, rapidly, and persistently interfering with gene expression in vivo. It is widely used in gene function research, drug target screening, cell signaling pathway analysis, and disease treatment.
1. Direct expression of shRNA, as shown in Figure 2, taking AAV virus vector as an example: the expression of shRNA is directly initiated by the type III promoter (U6 or H1), with strong expression ability and wide expression range, suitable for broad-spectrum expression;
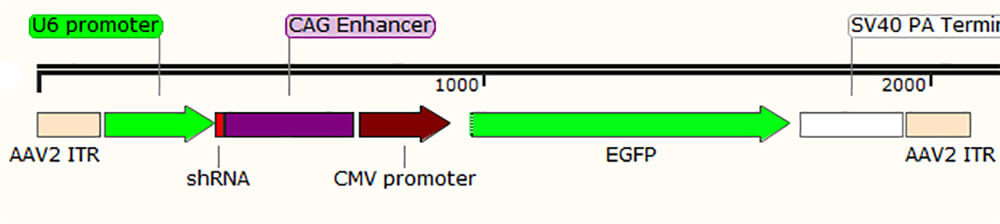
Figure 2. Key elements of rAAV-direct expression shRNA vector
2.Cre depends on shRNA, as shown in Figure 3, taking AAV virus vector as an example: Cre recombinase regulates the type III promoter (U6 or H1) to start the expression of shRNA, and the expression specificity is regulated by the expression of Cre, so it needs to be used with Cre virus or Cre mice. It has the advantages of specific expression and regulation by Cre expression, but its working efficiency is affected by the strength of Cre expression;
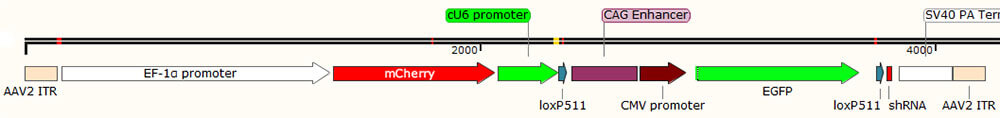
Figure 3. Key components of rAAV-Cre-dependent shRNA expression vector
3.miRNA-dependent shRNA, as shown in Figure 4, taking the AAV virus vector as an example: the system that expresses shRNA with miRNA as the backbone is promoted by the type II promoter, has strong expression ability and good interference effect, and the specificity of the type II promoter can be used to promote the specific expression of shRNA.

Figure 4. Key elements of rAAV-miRNA-dependent shRNA expression vector
| Category | No. | Product name | Tag |
| Direct expression | BC-0009 | pAAV-U6-shRNA(scramble)-CMV-EGFP-pA | EGFP |
| BC-0010 | pAAV-U6-shRNA(scramble)-CMV-mCherry-pA | mCherry | |
| Combined with Cre to achieve specific interference |
BC-0011 | pAAV-Efla-mCherry-U6-Loxp-CMV-EGFP-loxp-shRNA(scramble) | mCherry |
| pAAV-U6-loxp-CMV-EGFP-loxp-shRNA(scramble)-pA | EGFP | ||
| BC-0008 | pAAV-U6-loxp-CMV-mCherry-loxp-shRNA(scramble)-pA | mCherry | |
| miRNA-30a-specific interference | BC-0006 | pAAV-hSyn-EGFP-5'miR-30a-MCS-3'-miR30a-WPREs | EGFP |
| BC-0007 | pAAV-GFAP-EGFP-5'miR-30a-MCS-3'-miR30a-WPREs | EGFP | |
| In addition to providing coding gene shRNA interference services, Braincase also provides non-coding gene circRNA, lncRNA, MicroRNA interference services. | |||
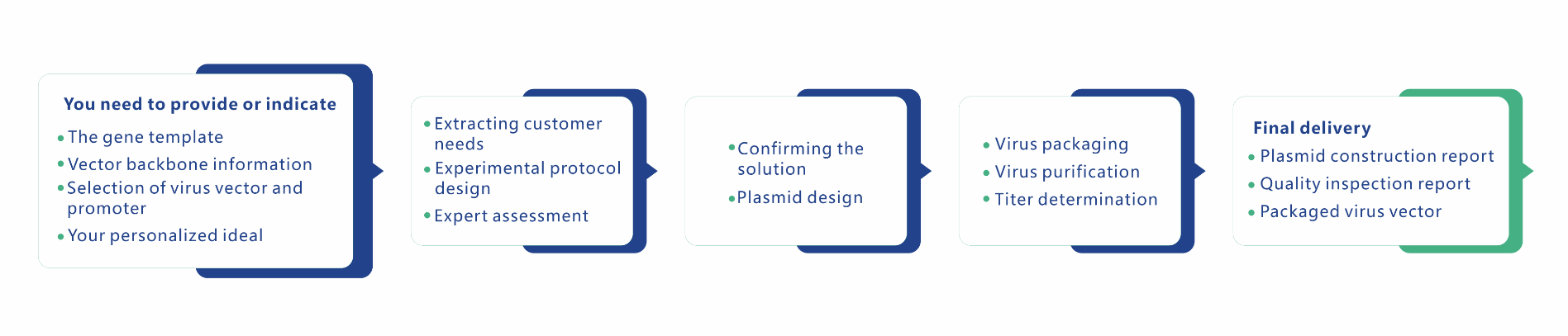
Service Description
Please download the attchement to know more about service description.
Servicedescription.pdf203.79 KB