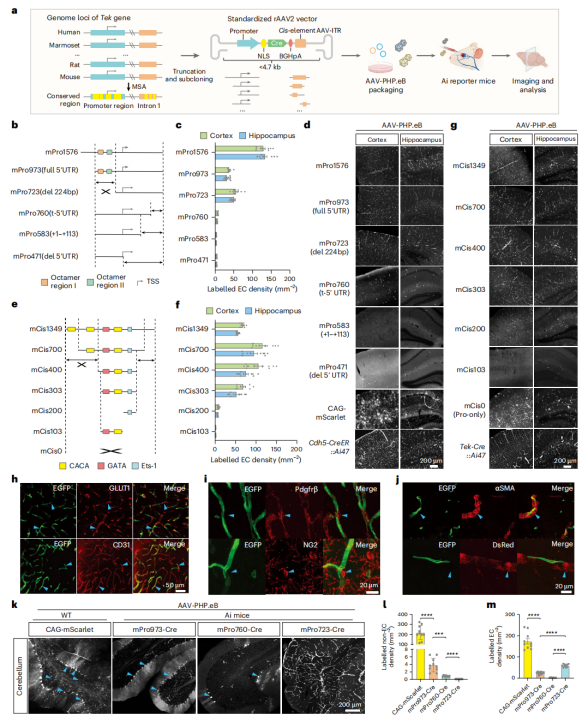
Construction of a bAVM Model Using the rAAV-miniBEND System
To establish a bAVM model using the rAAV-miniBEND system, the researchers selected the BrafV600E mutation—identified in bAVM patients but with unclear functional significance—as the target of investigation. AAV-PHP.eB-miniBEND-Cre was locally injected into the cortex or hippocampus of 30–50-day-old Braf-CAfl/fl mice, inducing the expression of the BRAFV600E mutant protein specifically in brain endothelial cells (Fig. 4a).
Within several weeks, the mice rapidly developed hallmark pathological features of bAVM, including intracerebral hemorrhage, edema, vascular malformations, and tissue necrosis. Additionally, the model mice exhibited hemiplegia and seizures, with a 3-month survival rate below 50% (Fig. 4c–f).
TUNEL staining, vascular labeling, and in vivo imaging revealed that bAVM lesions were dominated by abnormally dilated and malformed vessels, lacking normal capillary structures. The vessel diameter progressively increased over time, leading to altered local hemodynamics (Fig. 4g–l).
Two-photon imaging further demonstrated blood–brain barrier disruption (evidenced by plasma leakage) and the presence of endothelial-to-mesenchymal transition (EndMT) cell clusters, suggesting that EndMT may underlie the cellular basis of bAVM formation (Fig. 4j–n).
Finally, latex perfusion and PEGASOS tissue clearing imaging directly confirmed the occurrence of arteriovenous shunting in the brain—representing the core pathological hallmark of bAVM (Fig. 4o,p).
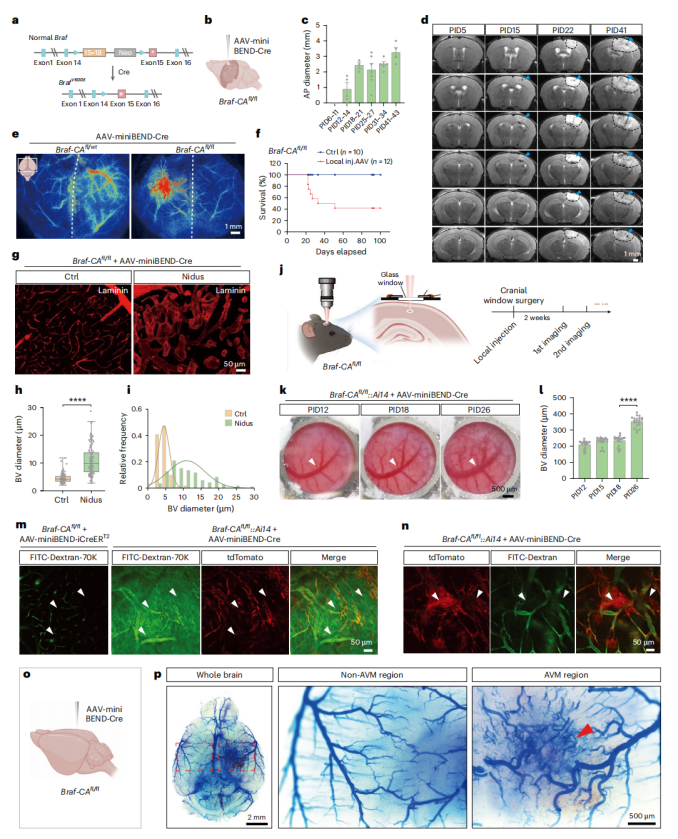 Figure 4 | Modeling bAVM using the rAAV-miniBEND system.
Figure 4 | Modeling bAVM using the rAAV-miniBEND system.
To evaluate the therapeutic potential of this model, the researchers intervened with the BRAFV600E inhibitor PLX4032 (vemurafenib) after bAVM induction. The results showed that, compared with the control group, drug treatment significantly reduced the lesion size and the density and diameter of malformed vessels (Fig. 5a–g). This indicates that activation of BRAF and its downstream pathways drives bAVM pathogenesis.
In summary, the rAAV-miniBEND system successfully established a BrafV600E-mediated bAVM mouse model, providing a new platform for cerebrovascular malformation mechanism studies and drug screening.
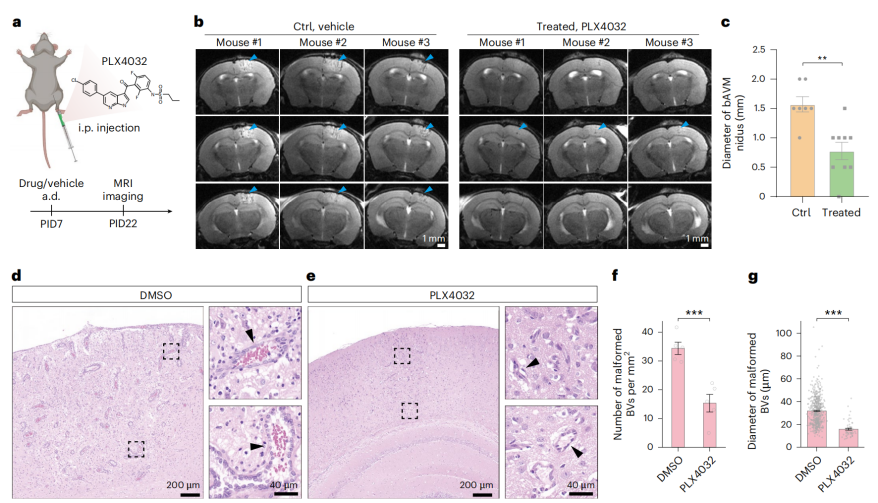
Figure 5 | PLX4032 treatment for bAVM.
Mechanistic Study of bAVM Development and Progression
To elucidate the molecular mechanisms underlying bAVM, researchers performed single-nucleus RNA sequencing on lesioned tissues 15 days (early stage) and 27 days (late stage) after local injection of AAV-miniBEND-Cre virus into the cortex of Braf-CAAᶠˡ/ᶠˡ mice. Cluster analysis revealed several stage-specific cell populations in the lesions (Fig. 6a–c), accompanied by a series of differentially expressed genes — such as downregulation of Slco1a4 and Spock2, and upregulation of Tll1 and Ackr1. These transcriptional alterations showed strong consistency with endothelial transcriptome profiles from human bAVM patients (Fig. 6d–i, 7b).
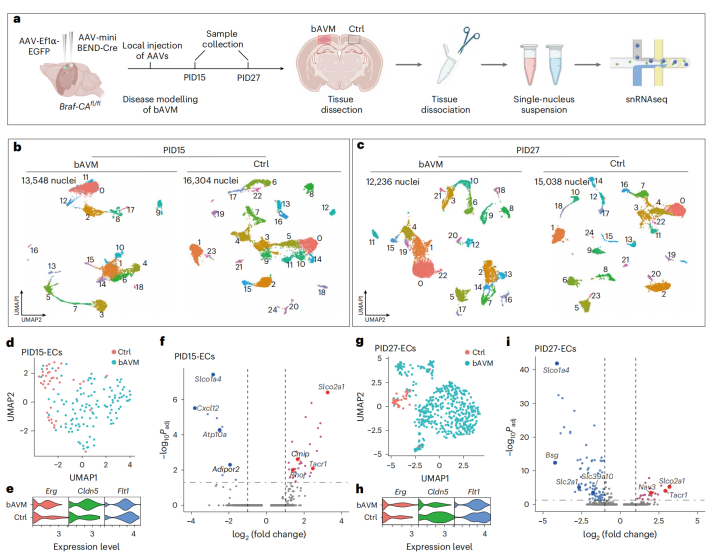
Figure 6. Single-nucleus RNA sequencing (snRNA-seq) reveals the molecular mechanisms underlying the formation and progression of bAVM.
Gene Ontology (GO) enrichment analysis demonstrated that genes upregulated in both Braf-bAVM mice and human patients were significantly enriched in angiogenesis and epithelial/endothelial cell migration processes (Fig. 7a). Moreover, proliferation-related genes (Mki67, Cdk1, Cdk6, Ccnd3, Gse1, and Nrp2) were markedly upregulated in endothelial cells within the lesions (Fig. 7b), and Ki67 staining confirmed a notable increase in proliferating cells in the affected regions (Fig. 7c–g). Both tip cell markers (Flt4, Dll4, Robo4, Kdr, Nrp2, and Nrp1) and stalk cell markers (Notch1, Jag1, and Rhoa) were upregulated (Fig. 7h–i), indicating the presence of a pathologically hyperactive angiogenic process driven by the BRAFV600E mutation.
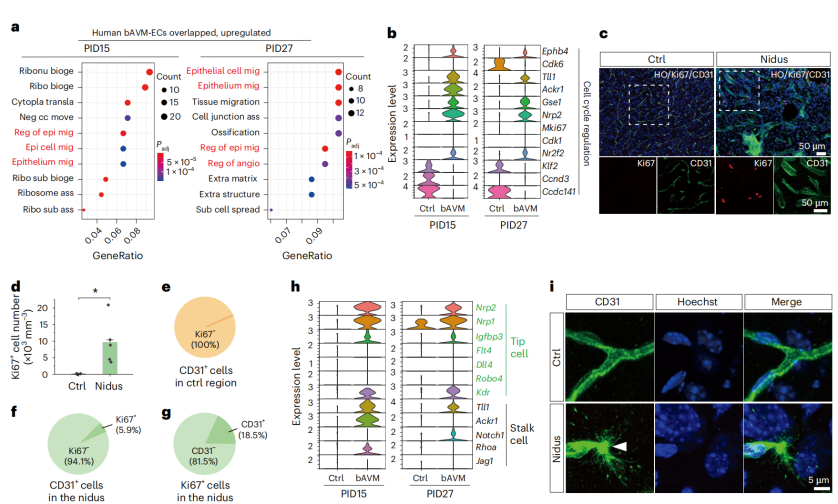
Figure 7. Identification of key signaling pathways and genes involved in bAVM onset and progression.
Further, single-cell RNA sequencing of cortical endothelial cells following AAV-miniBEND-Cre treatment (Fig. 8a) yielded 5,646 endothelial cells (2,624 from the bAVM group and 3,022 from controls). Clustering analysis identified three endothelial subpopulations — arterial, venous, and capillary (Fig. 8b–d). Notably, the proportion of venous endothelial cells (vECs) was significantly increased in the bAVM group (Fig. 8e–g), with differentially expressed genes mainly associated with vascular inflammation, endothelial proliferation, and dysfunction (Fig. 8h). Gene Set Enrichment Analysis (GSEA) revealed that angiogenesis-related and ERK1/2 signaling pathways were strongly activated in vECs (Fig. 8i–l), highlighting their critical role in bAVM progression.
In summary, the rAAV-miniBEND–mediated BrafV600E model successfully recapitulates the key molecular features of human bAVM and reveals the central role of venous endothelial cells in its formation and progression.
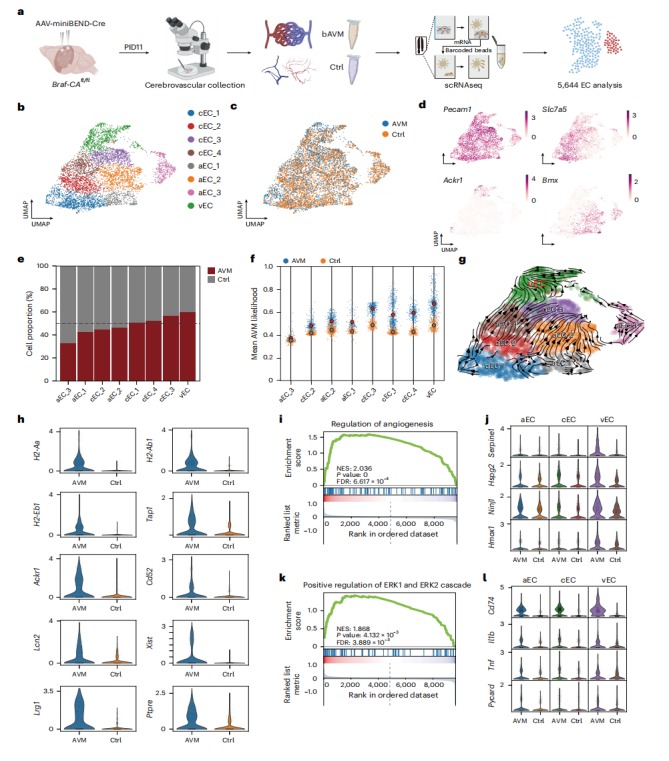 Figure 8. Single-cell RNA sequencing analysis of brain endothelial cells in BrafV600E-induced AVM.
Figure 8. Single-cell RNA sequencing analysis of brain endothelial cells in BrafV600E-induced AVM.
Summary
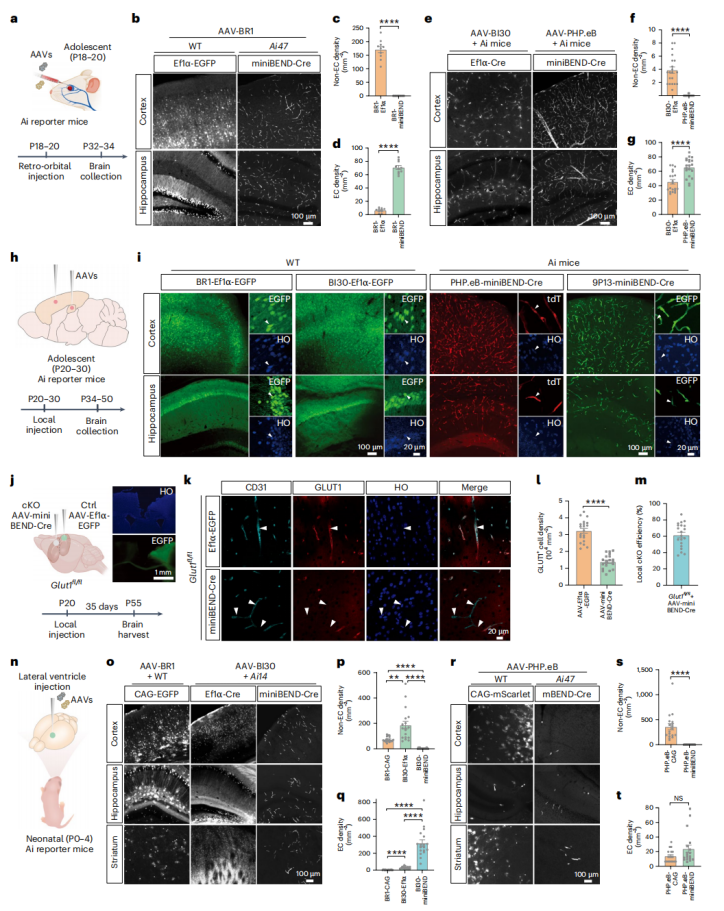
In this study, the rAAV-miniBEND system was successfully developed through optimization of Tek gene regulatory elements, achieving efficient and specific gene delivery to brain endothelial cells with a large payload capacity of 2.8 kb. The system supports multiple administration routes (intracranial or systemic injection) and enables highly specific targeting of brain endothelial cells at different developmental stages in both mice and rats, while effectively minimizing off-target effects in peripheral organs.
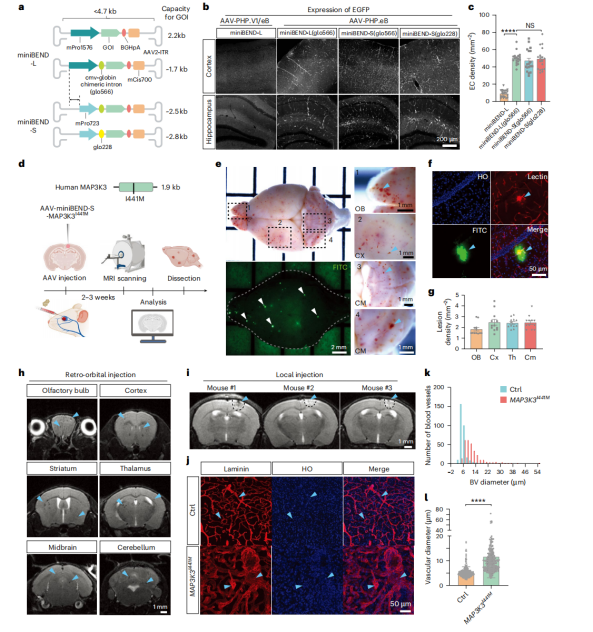
Using rAAV-miniBEND, researchers successfully generated a MAP3K3I441M-mediated CCM model and a BrafV600E-mediated brain AVM model, which rapidly induce typical lesions within two weeks and allow for multi-modal real-time observation. This approach overcomes the limitations of traditional models, which are time-consuming and inefficient, providing a powerful experimental tool for cerebrovascular disease research and offering new opportunities for mechanistic studies and therapeutic development of cerebrovascular malformations.
The rAAV-miniBEND viral tools used in this study are available from
Brain Case Biotech(bd@ebraincase.com)