- E-mail:BD@ebraincase.com
- Tel:+8618971215294
As neuroscience research advances into a multidimensional era of “cell type–circuit–behavior” analysis, traditional neuron classification methods based on antibody labeling face several limitations—such as insufficient antibody specificity, high cost, and low throughput (e.g., difficulty labeling multiple proteins on the same brain slice). These challenges significantly hinder the in-depth investigation of complex neural circuits and disease mechanisms.
To overcome these barriers, we have launched an innovative research service: “Neural Circuit Tracing + Targeted Spatial Omics + Neuronal Classification Analysis.” After performing neural circuit tracing, we apply spatial transcriptomics to the brain tissue containing the labeled neurons. This approach enables the integration of anatomical connectivity data from tracing with spatially resolved gene expression profiles.
Such integration allows researchers to directly associate labeled neurons with their molecular characteristics—such as expression levels of specific genes or neuronal subtypes—thereby uncovering direct links between neuron types, circuit connectivity, and functional states.
This service is built upon three core technologies:
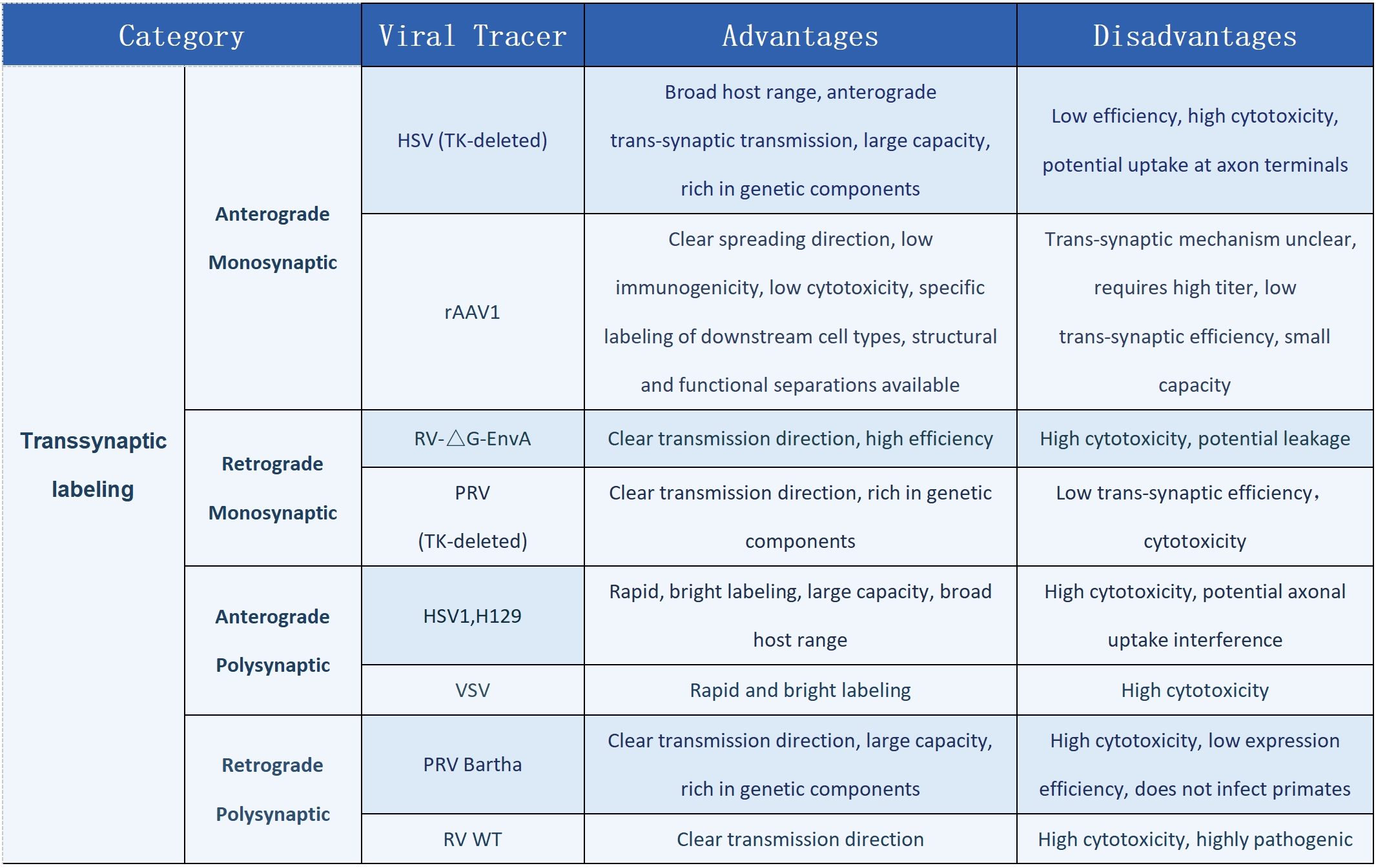
Utilizing viral tools such as AAV, RV, and PRV, this technique enables anterograde/retrograde and trans-synaptic labeling, allowing precise mapping of neuronal projection pathways.
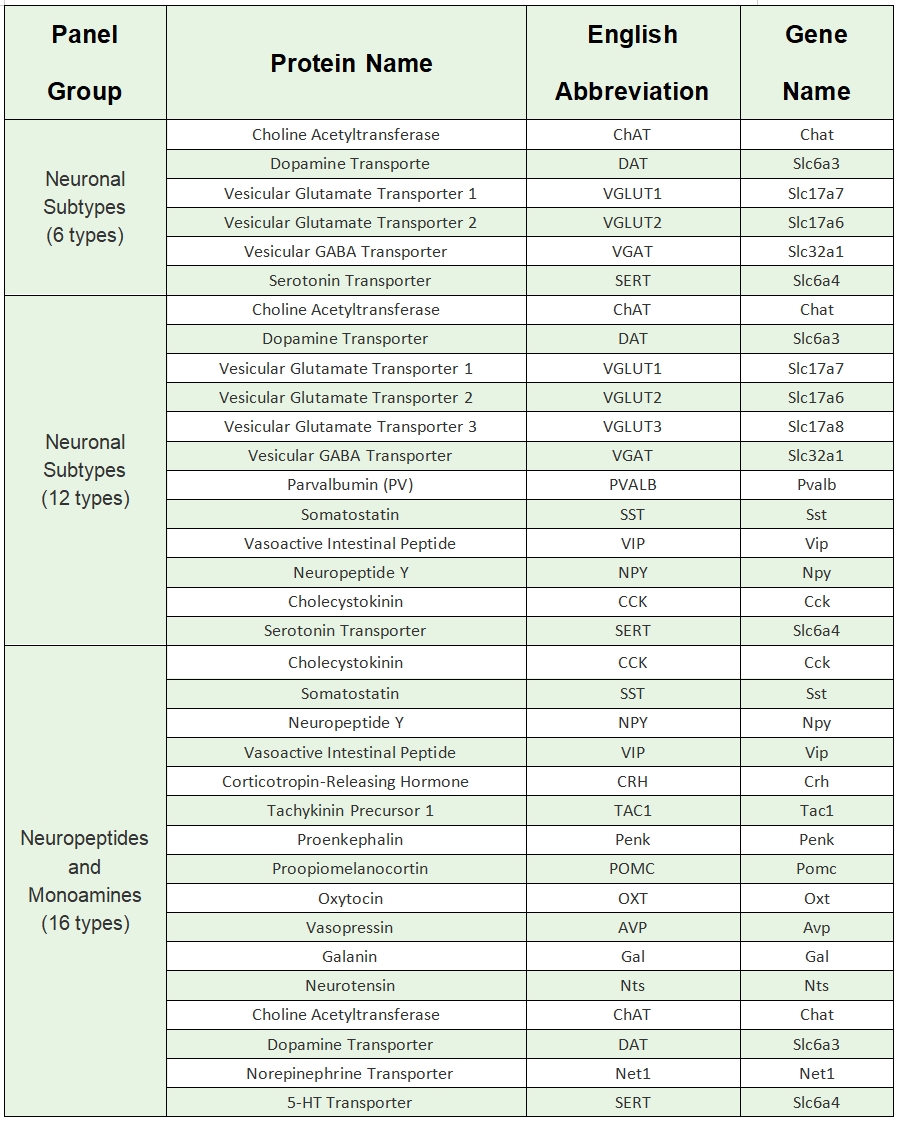
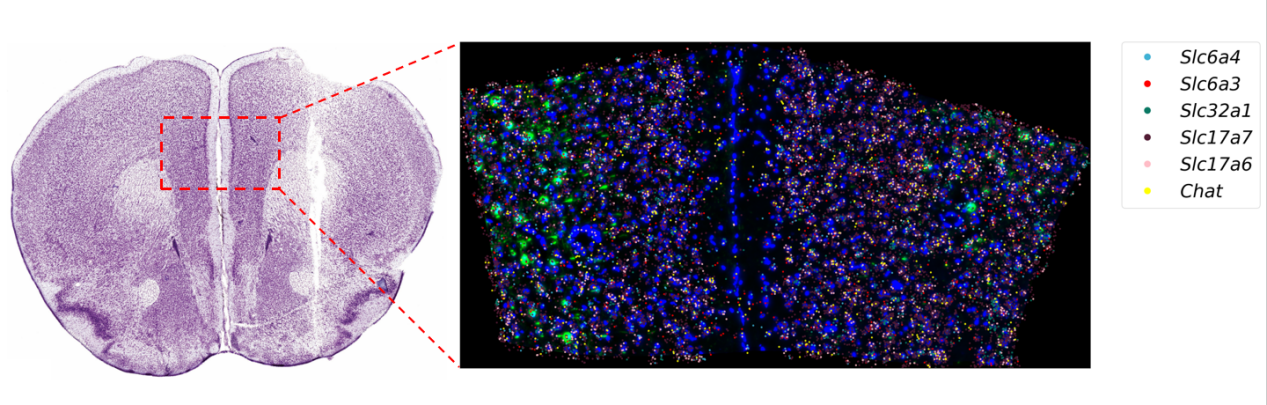
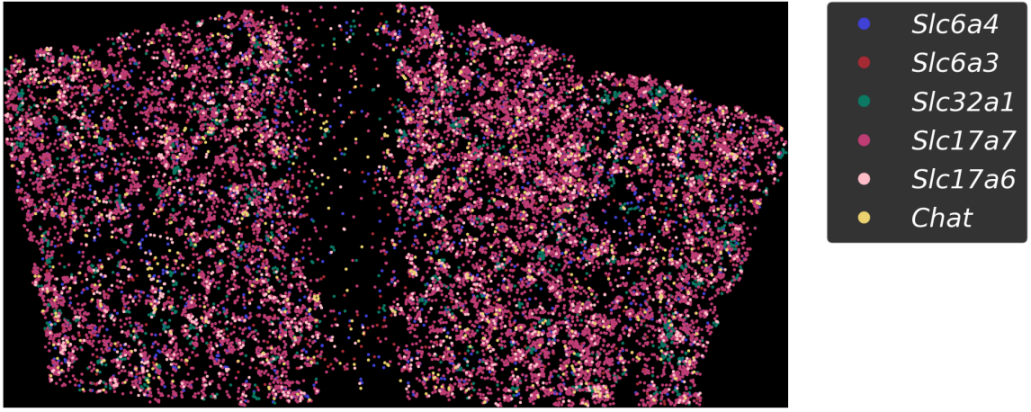
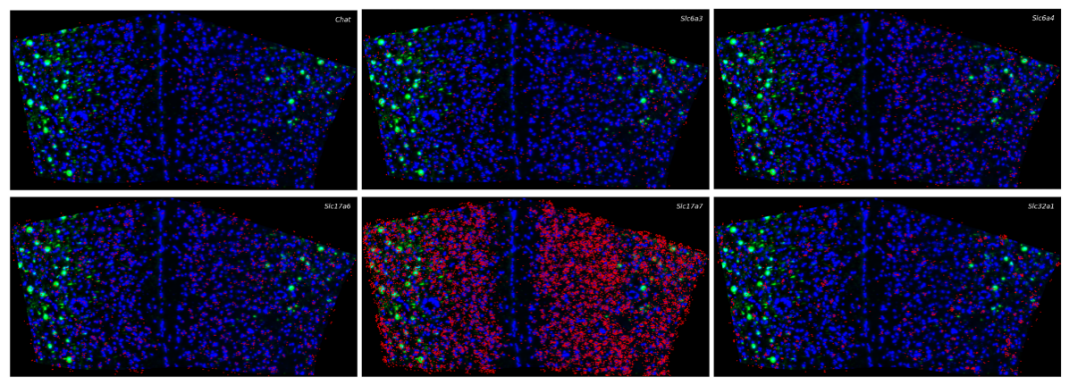
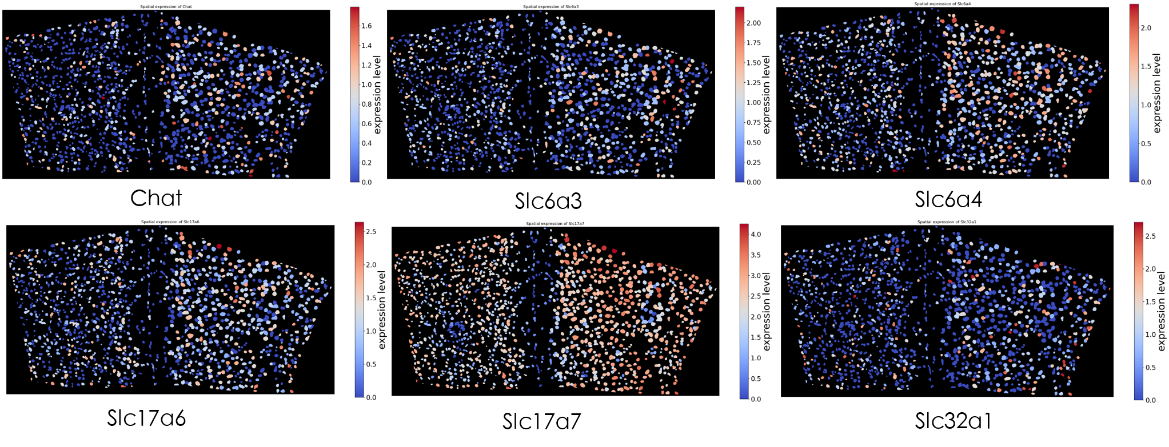
A novel targeted spatial omics technology featuring high sensitivity, high specificity, and high resolution. It enables qualitative and quantitative detection at single-base and subcellular levels, allowing the construction of single-cell spatial maps. MiP-seq is widely applied in fields such as neuroscience, development, and oncology.
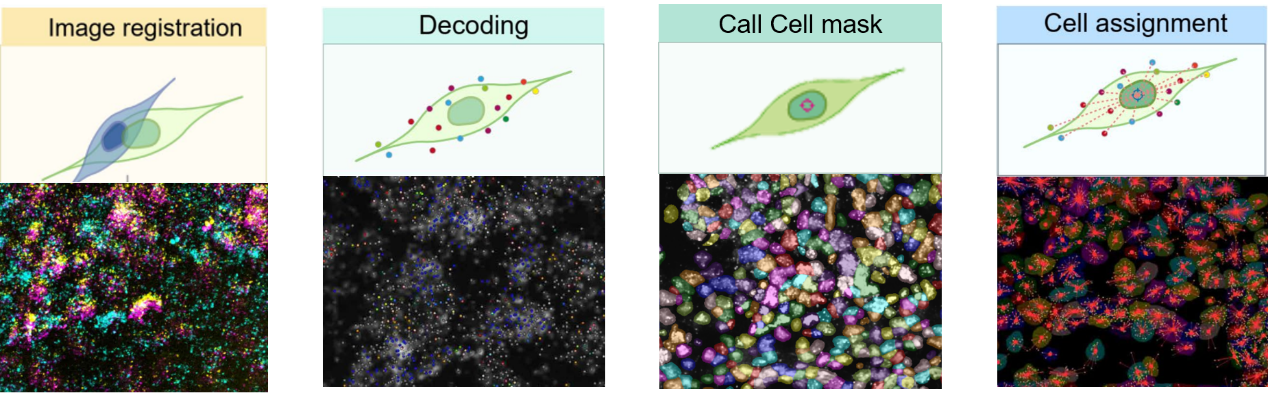
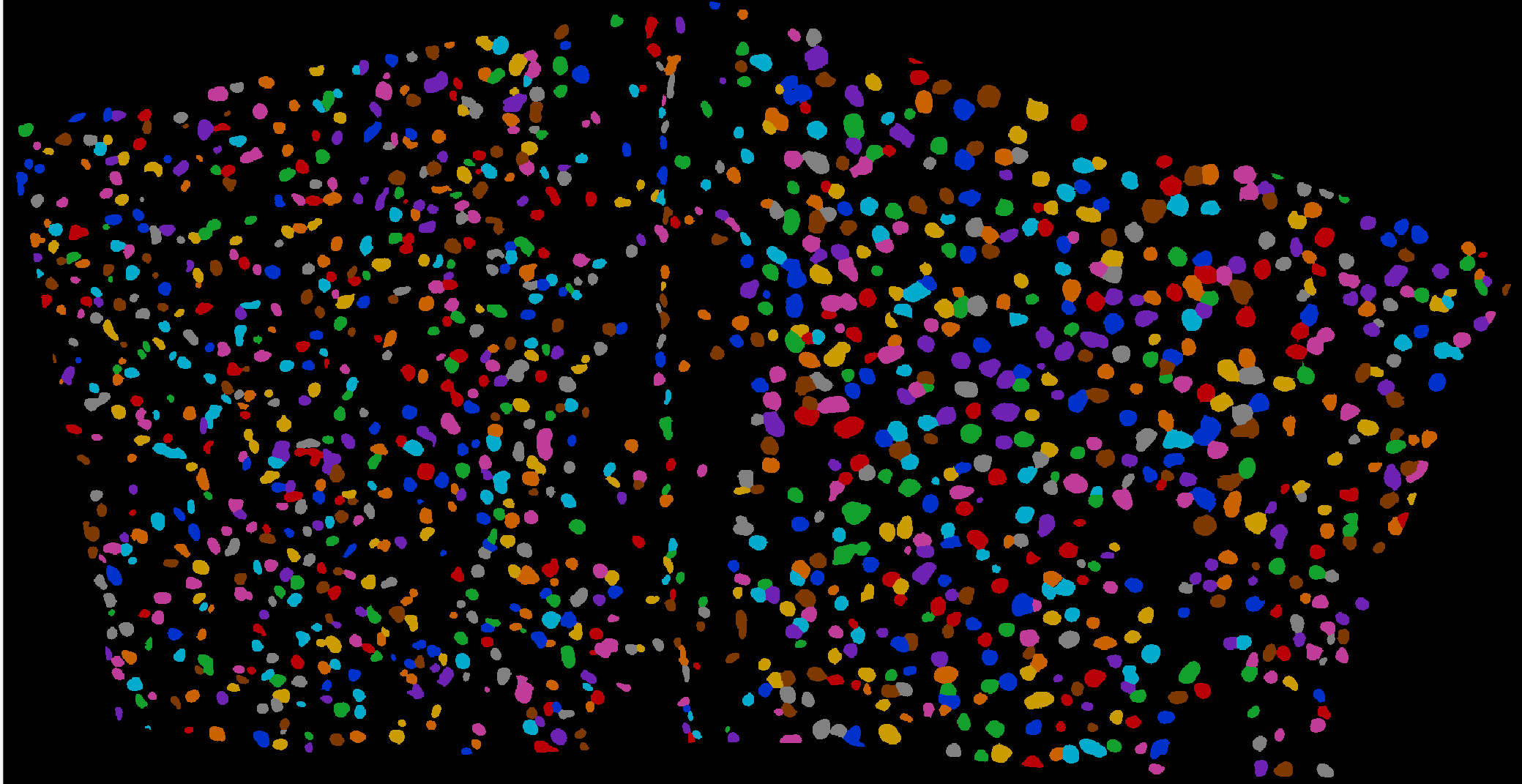
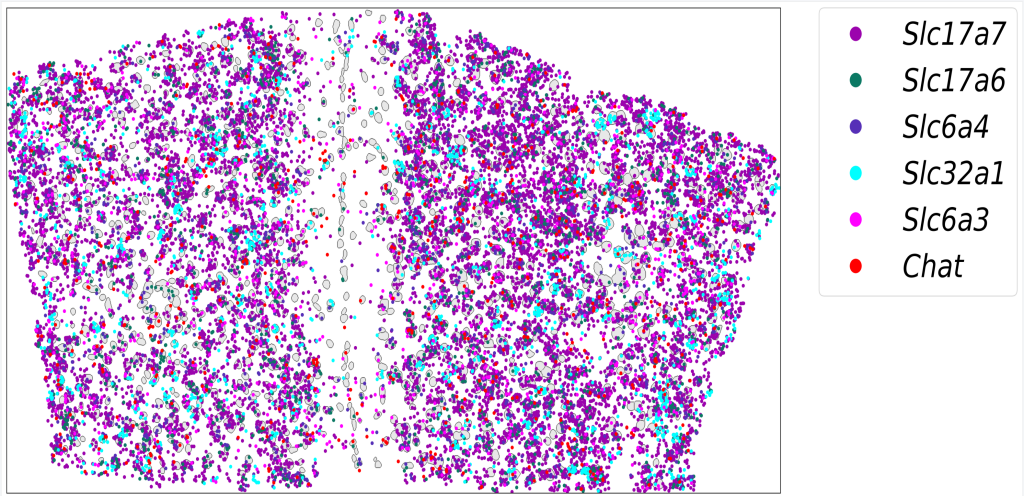
Integrating image processing, cell segmentation, gene expression quantification, and spatial localization to elucidate the relationships between neuron types and circuit functions.